WFMM
WFMM is a Windows command-line application that implements a Bayesian
wavelet-based functional mixed model methodology for functional data analysis
introduced in Morris and Carroll (2006).
The method extends linear mixed models to functional data consisting of n
curves sampled on the same grid. The user provides a file in Matlab data file format (*.mat) containing a matrix of data
samples of the set of n curves sampled T times and a description of the model
by the design matrix X and random effects matrix Z, and other parameters
controlling the computation. It provides as output nonparametric estimates of
fixed and random effects functions that have been adaptively regularized as a
result of the nonlinear shrinkage prior imposed on the fixed effects wavelet
coefficients.
See the WFMM User Guide, also included in
the download distribution. Current users of version 3.0 should read Changes to WFMM in Version 3.1
for new features in version 3.1 and some things to watch out for when moving from 3.0 to 3.1.
We strongly recommend that new users download the example file and run the
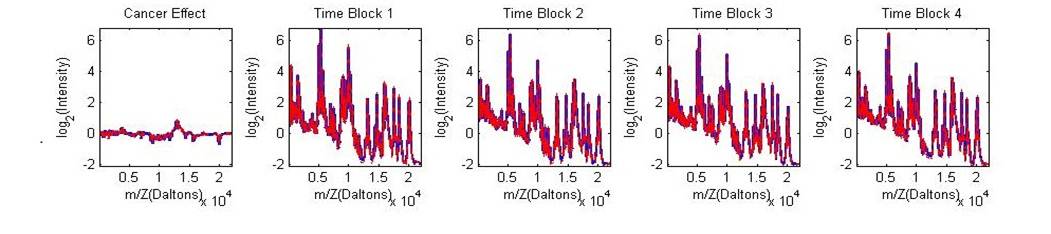
example data set. This is a partial spectrum of a MALDI-TOF mass spectrometry
proteomic data set from the pancreatic cancer experiment described in Morris et
al (2008). The plots below show results for the five fixed effects (cancer
effect and four block effects).? Open the
included Pancreatic_MYO25_wfmm_example.fig file in Matlab
and use the zoom to see the individual curves. The blue line is beta_ns (non-shrunken estimate for fixed effect), the solid
red line is beta_mean (posterior mean of fixed
effect), and the two red dashed lines are beta_quantiles? where 0.05 and 0.95 have been
specified.
The example requires Matlab to be installed on
your system. After download, unzip the files into a folder,
open a command prompt window in the folder and type
wfmmdemo.bat
The batch file runs wfmm,
then starts Matlab to plot
the results, which should look like the figure above. If Matlab does not
run successfully, start it manually, change current folder to the present
directory and run the PlotPancreatic_MYO25.m script.
Software developed in C++ and Matlab by Richard
Herrick, Ph.D.
References
Morris, JS and Carroll, RJ (2006). Wavelet-based functional mixed models, Journal of the Royal Statistical Society, Series
B, 68(2): 179-199
Morris JS,
Arroyo C, Coull B, Ryan LM, Herrick R, and Gortmaker SL (2006). Using Wavelet-Based Functional Mixed Models to characterize Population Heterogeneity in Accelerometer Profiles: A Case Study. Journal of the
American Statistical Association, 101(476): 1352-1364
Morris, JS, Brown PJ, Herrick, RC, Baggerly
KA, and Coombes, KR (2008). Bayesian Analysis of Mass Spectrometry Proteomic Data using Wavelet Based Functional Mixed Models,
Biometrics 64(2): 479-489.
Morris , JS, Baladandayuthapani,
V, Herrick, RC, Sanna, P, and Gutstein,
H (2011). Automated Analysis of Quantitative Image Data Using Isomorphic Functional Mixed Models, with Applications to Proteomics Data, The
Annals of Applied Statistics, 5(2A), 894-923.
 Biostatistics Software --- Desktop / Cloud
Biostatistics Software --- Desktop / Cloud
